from bioMONAI.data import *
from bioMONAI.transforms import *
from bioMONAI.core import *
from bioMONAI.core import Path
from bioMONAI.data import get_image_files, get_target, RandomSplitter
from bioMONAI.nets import BasicUNet, DynUNet
from bioMONAI.losses import *
from bioMONAI.metrics import *
from bioMONAI.datasets import download_medmnist
from monai.utils import set_determinism
set_determinism(0)Tutorial Classification
Tutorial Classification
device = get_device()
print(device)cpuCUDA initialization: CUDA unknown error - this may be due to an incorrectly set up environment, e.g. changing env variable CUDA_VISIBLE_DEVICES after program start. Setting the available devices to be zero. (Triggered internally at /opt/conda/conda-bld/pytorch_1682343995622/work/c10/cuda/CUDAFunctions.cpp:109.)Download Data
image_path = '../_data/medmnist_data/'
info = download_medmnist('bloodmnist', image_path, download_only=True)
infoDownloading https://zenodo.org/records/10519652/files/bloodmnist.npz?download=1 to ../_data/medmnist_data/bloodmnist.npz
Using downloaded and verified file: ../_data/medmnist_data/bloodmnist.npz
Using downloaded and verified file: ../_data/medmnist_data/bloodmnist.npz
Saving training images to ../_data/medmnist_data/...
Saving validation images to ../_data/medmnist_data/...
Saving test images to ../_data/medmnist_data/...
Removed bloodmnist.npz
Datasets downloaded to ../_data/medmnist_data/
Dataset info for 'bloodmnist': {'python_class': 'BloodMNIST', 'description': 'The BloodMNIST is based on a dataset of individual normal cells, captured from individuals without infection, hematologic or oncologic disease and free of any pharmacologic treatment at the moment of blood collection. It contains a total of 17,092 images and is organized into 8 classes. We split the source dataset with a ratio of 7:1:2 into training, validation and test set. The source images with resolution 3×360×363 pixels are center-cropped into 3×200×200, and then resized into 3×28×28.', 'url': 'https://zenodo.org/records/10519652/files/bloodmnist.npz?download=1', 'MD5': '7053d0359d879ad8a5505303e11de1dc', 'url_64': 'https://zenodo.org/records/10519652/files/bloodmnist_64.npz?download=1', 'MD5_64': '2b94928a2ae4916078ca51e05b6b800b', 'url_128': 'https://zenodo.org/records/10519652/files/bloodmnist_128.npz?download=1', 'MD5_128': 'adace1e0ed228fccda1f39692059dd4c', 'url_224': 'https://zenodo.org/records/10519652/files/bloodmnist_224.npz?download=1', 'MD5_224': 'b718ff6835fcbdb22ba9eacccd7b2601', 'task': 'multi-class', 'label': {'0': 'basophil', '1': 'eosinophil', '2': 'erythroblast', '3': 'immature granulocytes(myelocytes, metamyelocytes and promyelocytes)', '4': 'lymphocyte', '5': 'monocyte', '6': 'neutrophil', '7': 'platelet'}, 'n_channels': 3, 'n_samples': {'train': 11959, 'val': 1712, 'test': 3421}, 'license': 'CC BY 4.0'}100%|██████████| 35461855/35461855 [00:11<00:00, 3024955.63it/s]
100%|██████████| 11959/11959 [00:02<00:00, 5592.09it/s]
100%|██████████| 1712/1712 [00:00<00:00, 5460.00it/s]
100%|██████████| 3421/3421 [00:00<00:00, 5433.55it/s]{'python_class': 'BloodMNIST',
'description': 'The BloodMNIST is based on a dataset of individual normal cells, captured from individuals without infection, hematologic or oncologic disease and free of any pharmacologic treatment at the moment of blood collection. It contains a total of 17,092 images and is organized into 8 classes. We split the source dataset with a ratio of 7:1:2 into training, validation and test set. The source images with resolution 3×360×363 pixels are center-cropped into 3×200×200, and then resized into 3×28×28.',
'url': 'https://zenodo.org/records/10519652/files/bloodmnist.npz?download=1',
'MD5': '7053d0359d879ad8a5505303e11de1dc',
'url_64': 'https://zenodo.org/records/10519652/files/bloodmnist_64.npz?download=1',
'MD5_64': '2b94928a2ae4916078ca51e05b6b800b',
'url_128': 'https://zenodo.org/records/10519652/files/bloodmnist_128.npz?download=1',
'MD5_128': 'adace1e0ed228fccda1f39692059dd4c',
'url_224': 'https://zenodo.org/records/10519652/files/bloodmnist_224.npz?download=1',
'MD5_224': 'b718ff6835fcbdb22ba9eacccd7b2601',
'task': 'multi-class',
'label': {'0': 'basophil',
'1': 'eosinophil',
'2': 'erythroblast',
'3': 'immature granulocytes(myelocytes, metamyelocytes and promyelocytes)',
'4': 'lymphocyte',
'5': 'monocyte',
'6': 'neutrophil',
'7': 'platelet'},
'n_channels': 3,
'n_samples': {'train': 11959, 'val': 1712, 'test': 3421},
'license': 'CC BY 4.0'}Create Dataloader
from fastai.vision.all import *bs, size = 4, 128
path = Path(image_path)
path_train = path/'train'
path_val = path/'val'
data_ops = {
'blocks': (BioImageBlock(cls=BioImageMulti), CategoryBlock(vocab=info['label'])),
'get_items': get_image_files,
'get_y': parent_label,
'splitter': GrandparentSplitter(),
'item_tfms': [ScaleIntensity(min=0.0, max=1.0),RandCrop2D(size), RandRot90(prob=0.5), RandFlip(prob=0.75)],
'bs': bs,
}
data = get_dataloader(
path,
show_summary=True,
**data_ops,
)
# print length of training and validation datasets
print('train images:', len(data.train_ds.items), '\nvalidation images:', len(data.valid_ds.items))Setting-up type transforms pipelines
Collecting items from ../_data/medmnist_data
Found 17092 items
2 datasets of sizes 13674,3418
Setting up Pipeline: BioImageMulti.create -> Tensor2BioImage -- {}
Setting up Pipeline: parent_label -> Categorize -- {'vocab': ['0', '1', '2', '3', '4', '5', '6', '7'], 'sort': True, 'add_na': False}
Building one sample
Pipeline: BioImageMulti.create -> Tensor2BioImage -- {}
starting from
../_data/medmnist_data/train/5/train_2282.png
applying BioImageMulti.create gives
BioImageMulti of size 3x28x28
applying Tensor2BioImage -- {} gives
BioImageMulti of size 3x28x28
Pipeline: parent_label -> Categorize -- {'vocab': ['0', '1', '2', '3', '4', '5', '6', '7'], 'sort': True, 'add_na': False}
starting from
../_data/medmnist_data/train/5/train_2282.png
applying parent_label gives
5
applying Categorize -- {'vocab': ['0', '1', '2', '3', '4', '5', '6', '7'], 'sort': True, 'add_na': False} gives
TensorCategory(5)
Final sample: (BioImageMulti([[[209., 222., 224., ..., 252., 253., 253.],
[219., 211., 210., ..., 252., 253., 253.],
[212., 211., 219., ..., 252., 253., 253.],
...,
[255., 255., 255., ..., 199., 215., 231.],
[251., 255., 255., ..., 212., 232., 242.],
[250., 255., 255., ..., 223., 242., 250.]],
[[171., 187., 189., ..., 232., 231., 231.],
[184., 177., 176., ..., 232., 231., 231.],
[179., 178., 186., ..., 232., 231., 231.],
...,
[230., 233., 232., ..., 176., 194., 210.],
[228., 233., 233., ..., 190., 210., 221.],
[228., 233., 235., ..., 200., 221., 228.]],
[[182., 194., 193., ..., 207., 208., 207.],
[188., 178., 175., ..., 207., 207., 207.],
[172., 171., 179., ..., 205., 207., 208.],
...,
[190., 196., 197., ..., 184., 189., 193.],
[187., 195., 195., ..., 192., 199., 200.],
[187., 194., 196., ..., 194., 204., 204.]]]), TensorCategory(5))
Collecting items from ../_data/medmnist_data
Found 17092 items
2 datasets of sizes 13674,3418
Setting up Pipeline: BioImageMulti.create -> Tensor2BioImage -- {}
Setting up Pipeline: parent_label -> Categorize -- {'vocab': ['0', '1', '2', '3', '4', '5', '6', '7'], 'sort': True, 'add_na': False}
Setting up after_item: Pipeline: ScaleIntensity -> RandCrop2D -- {'size': (128, 128), 'lazy': False, 'p': 1.0} -> RandRot90 -- {'prob': 0.5, 'max_k': 3, 'spatial_axes': (0, 1), 'ndim': 2, 'lazy': False, 'p': 1.0} -> RandFlip -- {'prob': 0.75, 'spatial_axis': None, 'ndim': 2, 'lazy': False, 'p': 1.0} -> ToTensor
Setting up before_batch: Pipeline:
Setting up after_batch: Pipeline:
Building one batch
Applying item_tfms to the first sample:
Pipeline: ScaleIntensity -> RandCrop2D -- {'size': (128, 128), 'lazy': False, 'p': 1.0} -> RandRot90 -- {'prob': 0.5, 'max_k': 3, 'spatial_axes': (0, 1), 'ndim': 2, 'lazy': False, 'p': 1.0} -> RandFlip -- {'prob': 0.75, 'spatial_axis': None, 'ndim': 2, 'lazy': False, 'p': 1.0} -> ToTensor
starting from
(BioImageMulti of size 3x28x28, TensorCategory(5))
applying ScaleIntensity gives
(BioImageMulti of size 3x28x28, TensorCategory(5))
applying RandCrop2D -- {'size': (128, 128), 'lazy': False, 'p': 1.0} gives
(BioImageMulti of size 3x28x28, TensorCategory(5))
applying RandRot90 -- {'prob': 0.5, 'max_k': 3, 'spatial_axes': (0, 1), 'ndim': 2, 'lazy': False, 'p': 1.0} gives
(BioImageMulti of size 3x28x28, TensorCategory(5))
applying RandFlip -- {'prob': 0.75, 'spatial_axis': None, 'ndim': 2, 'lazy': False, 'p': 1.0} gives
(BioImageMulti of size 3x28x28, TensorCategory(5))
applying ToTensor gives
(BioImageMulti of size 3x28x28, TensorCategory(5))
Adding the next 3 samples
No before_batch transform to apply
Collating items in a batch
No batch_tfms to apply
None
train images: 13674
validation images: 3418data.show_batch(max_n=4)
Load and train a 2D model
# from monai.networks.nets import densenet121
# model = densenet121(spatial_dims=2, in_channels=3, out_channels=8)model = resnet18
loss = CrossEntropyLossFlat()
metrics = accuracy
trainer = visionTrainer(data, model, loss_fn=loss, metrics=metrics, show_summary=True)Sequential (Input shape: 4 x 3 x 28 x 28)
============================================================================
Layer (type) Output Shape Param # Trainable
============================================================================
4 x 64 x 14 x 14
Conv2d 9408 True
BatchNorm2d 128 True
ReLU
____________________________________________________________________________
4 x 64 x 7 x 7
MaxPool2d
Conv2d 36864 True
BatchNorm2d 128 True
ReLU
Conv2d 36864 True
BatchNorm2d 128 True
Conv2d 36864 True
BatchNorm2d 128 True
ReLU
Conv2d 36864 True
BatchNorm2d 128 True
____________________________________________________________________________
4 x 128 x 4 x 4
Conv2d 73728 True
BatchNorm2d 256 True
ReLU
Conv2d 147456 True
BatchNorm2d 256 True
Conv2d 8192 True
BatchNorm2d 256 True
Conv2d 147456 True
BatchNorm2d 256 True
ReLU
Conv2d 147456 True
BatchNorm2d 256 True
____________________________________________________________________________
4 x 256 x 2 x 2
Conv2d 294912 True
BatchNorm2d 512 True
ReLU
Conv2d 589824 True
BatchNorm2d 512 True
Conv2d 32768 True
BatchNorm2d 512 True
Conv2d 589824 True
BatchNorm2d 512 True
ReLU
Conv2d 589824 True
BatchNorm2d 512 True
____________________________________________________________________________
4 x 512 x 1 x 1
Conv2d 1179648 True
BatchNorm2d 1024 True
ReLU
Conv2d 2359296 True
BatchNorm2d 1024 True
Conv2d 131072 True
BatchNorm2d 1024 True
Conv2d 2359296 True
BatchNorm2d 1024 True
ReLU
Conv2d 2359296 True
BatchNorm2d 1024 True
AdaptiveAvgPool2d
AdaptiveMaxPool2d
____________________________________________________________________________
4 x 1024
Flatten
BatchNorm1d 2048 True
Dropout
____________________________________________________________________________
4 x 512
Linear 524288 True
ReLU
BatchNorm1d 1024 True
Dropout
____________________________________________________________________________
4 x 8
Linear 4096 True
____________________________________________________________________________
Total params: 11,707,968
Total trainable params: 11,707,968
Total non-trainable params: 0
Optimizer used: <function Adam>
Loss function: FlattenedLoss of CrossEntropyLoss()
Callbacks:
- TrainEvalCallback
- CastToTensor
- Recorder
- ProgressCallback
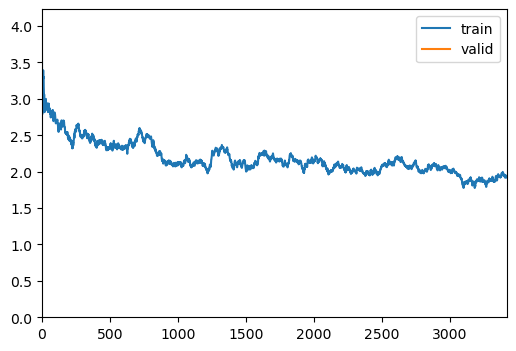
- ShowGraphCallbacktrainer.fit(1)| epoch | train_loss | valid_loss | accuracy | time |
|---|---|---|---|---|
| 0 | 1.941148 | 2.192013 | 0.252487 | 05:51 |
trainer.show_results(cmap='gray')



# trainer.save('tmp-model')Test data
Evaluate the performance of the selected model on unseen data. It’s important to not touch this data until you have fine tuned your model to get an unbiased evaluation!